Empowering Clinicians With Digitization and AI: A Nephrology Perspective
Authors:
Dr. phil. Aitana Neves
SIB Swiss Institute of Bioinformatics
Ch. des Mines 9, 1202 Genève
E-Mail: aitana.lebrand@sib.swiss
Dr. med. Ann-Kathrin Schwarzkopf
Zentrum für Urologie und Nephrologie
Hirslanden Bern AG
Salem-Spital
Schänzlistrasse 39, 3013 Bern
E-Mail: ann-kathrin.schwarzkopf@hin.ch
Artificial intelligence (AI) is gaining momentum in clinical care and translational clinical research. The concepts behind AI deserve to be demystified in order to ensure that those using these technologies, e.g. clinicians, understand their potential and also limitations. This is essential to build trust and set the ground to empower clinicians with e.g. novel applications or assistance in highly time-consuming tasks. We present several applications, published and envisioned, in the field of nephrology.
Keypoints
-
Machine learning (ML) is a concrete way of achieving artificial intelligence (AI) that allows identifying complex patterns from huge amounts of data.
-
The use of ML goes hand in hand with data digitization. Data used to train ML models should be previously well annotated and structured with defined variables and ontologies to facilitate its use and re-sharing.
-
An integrative, multidisciplinary approach is key: building trust by notably engaging early on with clinicians is essential for ML adoption.
-
The use of ML can support diagnosis, risk assessment, prognosis and treatment prediction with an emphasis on precision medicine (best available treatment based on individual risk profile).
Defining AI and Digitization
In recent years, artificial intelligence (AI) has gained momentum in all aspects of our lives. In fact, it is already present all around us, from recommender systems on online shops to chat bots on sites we visit, or for automated image recognition. In healthcare, AI has also made fast progress, notably in radiology, where it supports diagnosis and interpretation.1 We may compare the ongoing transformation to the conscious shift to evidence-based medicine, which has aimed to include clinical research and established professional guidelines into routine clinical decision making. In a nutshell, AI is the capacity of a computer to perform tasks that would typically require human intelligence. Machine learning (ML) is a concrete way of achieving AI, based on letting computers learn tasks from data. ML manages huge amounts of data and allows identifying complex patterns in the data thanks to elaborate models. This gives rise to data-driven medicine, which aims to integrate heterogeneous and large amounts of data to extract patterns that can then be used to support diagnosis, prognosis, risk assessment and treatment prediction. It basically uses the collective expertise hidden in existing data to an unprecedented level, taking into account many more variables than in more traditional evidence based medicine.
These AI models are driven by data, which need to be digitized to be used by computers. Digitization can occur in many forms, from scanning medical reports and images, to entering data into an electronic medical record (EMR), a laboratory information management system (LIMS) or a mobile health application (Fig.1). Quality and structure of the digitized data are important ingredients to train AI models aimed at supporting diagnosis, prognosis, risk assessment and treatment prediction. Quality refers to the fact that the data is as accurate and complete as possible. Data structure orders information into similar blocks, enabling the rapid identification of similar information and querying of the data. The example of a medical report scan would be coined unstructured data, because the information it contains is not saved into meaningful variables like patient name, symptoms, diagnosis, medication, etc. Structuring such data would require parsing the document to try to identify those elements in the free text, which can be achieved using an AI method called Text Mining. The resulting structured data could then be used further by other AI methodologies to support e.g. treatment prediction. EMR and LIMS on the other hand structure data by design, by using different entry boxes for different variables, and by restricting the allowed terms for each variable using controlled vocabularies and ontologies like the International Classification of Diseases (ICD10).
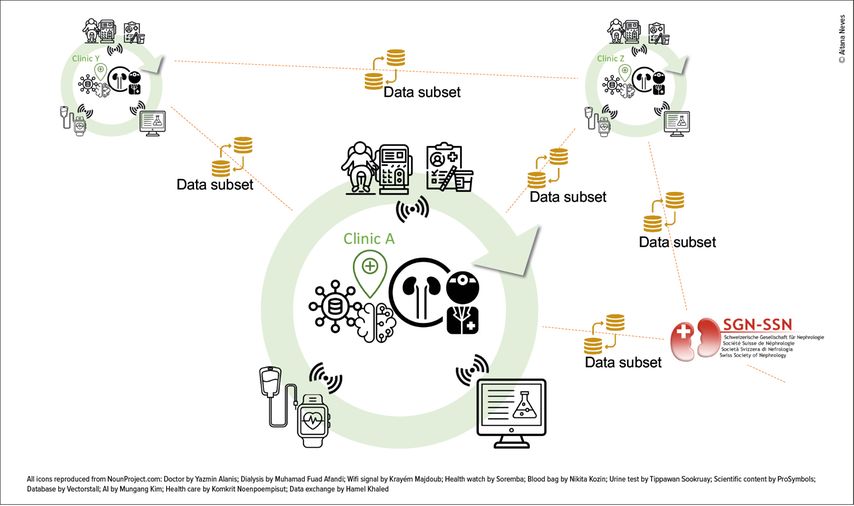
Fig. 1: Diverse data are generated and collected manually, semi-automatically or automatically, e.g. at the point of care (connected dialysis, sample collection etc.), at the laboratory that stores the results in a laboratory information management system or outside the clinic by connected devices (health wearables etc.) (green circles represent different clinics). In a digital ecosystem, all these data are digitized and made available to the physician e.g. through a clinical data warehouse interface. Digitized data can then be fueled into AI models to support diagnosis, prognosis, monitoring or treatment decisions. Parts of the data (in some cases anonymised) may also be shared between clinics or with national and international data repositories for better patient monitoring and quality of care, thanks to standardized and structured datasets (orange lines)
In this review, we explain the general concepts that underlie machine learning (ML) and what key elements are needed to fully explore its potential and understand its limitations with a focus in nephrology.2
How Digitization and ML May Serve Nephrology
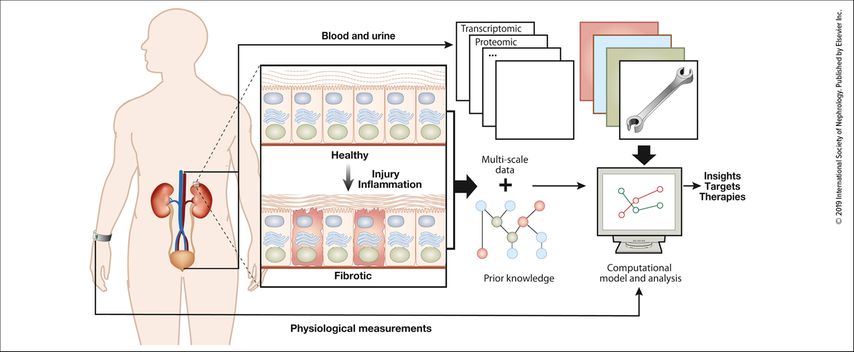
ML is already used in several fields of medicine and clinical translational research. Examples include image-based diagnostics disciplines such as radiology1 notably with computer-aided detection, and digital pathology.3 Integration of molecular data like DNA (genomics) and RNA (transcriptomics) thanks to next-generation sequencing, proteomics, biomarkers (metabolomics) and microbiome analyses creates even more complex big datasets.4 In this context, ML can be used for the annotation of these datasets. In oncology, molecular-based technologies (“omics”) and ML are already used to support diagnosis and predict response to a specific treatment.5 Precision medicine is the ambitious goal also in nephrology (Fig.2).6,7
Fig. 2: Overview of data generation and data integration from multiple sources (from Saez-Rodriguez J et al.: Kidney Int 2019; 95: 1326-37. Creative Commons CC-BY-NC-ND license)4
Coming to clinical nephrology, cohorts of interest are patients with the most common type of glomerulonephritis, IgA nephropathy,8 diabetic kidney disease (DKD) and acute kidney injury (AKI). As of now, therapeutic concepts in diabetes or other chronic kidney diseases rely on results from large clinical (randomized controlled) trials done in a group of patients. New treatments using SGLT2 inhibitors or mineralocorticoid receptor antagonists in diabetes are promising.9,10 But so far, despite lifestyle changes, optimizing blood glucose and blood pressure control as well as applying RAAS blockade, DKD is still the major factor causing end stage kidney disease ( www.usrds.org , www.era.org ). Genetic mutations have been associated with more aggressive kidney disease in either diabetes or IgA nephropathy and/or to respond to a specific therapy.8,11 Identifying these individual risk factors by computational analysis followed by closer surveillance and/or earlier intervention by risk stratification could improve individual outcome. To implement precision medicine in nephrology coming from one treatment for all to personalized diagnosis and personalized treatment, the kidney precision medicine project was founded ( www.kpmp.org ).
Compared to chronic kidney disease, AKI is an increasingly frequent condition that generates adverse events and high costs. To reduce associated high morbidity and mortality in this heterogeneous group, the American Society of Nephrology has established the AKI!now Initiative to improve outcomes or even better to prevent AKI.12 ML has been used to develop prognostic bedside tools predicting postoperative or sepsis-associated AKI ( www.akipredictor.com/en ).13,14
Finally in the field of kidney transplantation, AI studies are focused on radiology evaluation, molecular diagnostics of tissue, prediction of early graft function and survival, diagnosis of rejection and optimizing dose of immunosuppression.15
Essential Ingredients to Build Robust and Trustworthy ML Models
Getting High Quality Data
ML models rely on annotated data for learning a task. For example, to predict if a patient is at risk of AKI, it will require data of patients annotated as low, medium and high risks. This annotation by experts is where the collective expertise is fed into the model. It tells the model what the expected prediction should be, so it can learn. If this annotation step is of low quality, we end up with garbage in, garbage out. Data-driven models rely on data and its quality will determine the overall quality of the predictions.
Choosing the Model
Given a training dataset properly annotated, the model can start learning. In this training process, the importance of all the input variables will be gradually adjusted, until the model prediction matches the annotation. Once the importance of each variable is set, the model can be used to make predictions on new data. Most of the expertise in ML relies on choosing the right type of model for the data and task at hand, and ensuring that this optimization process, called training, is done properly. A model is a mathematical representation of the relationship between the input variables and the target output. The most simple model would be a multivariate linear regression where the number of parameters is of the order of the number of input variables. ML actually relies on much more sophisticated models that also enable extracting complex non-linear patterns from the data. Examples of commonly used models are random forests and deep learning,16 which is a particular type of artificial neural network (ANN).
Building Trust
Ultimately, the choice of the task to solve, of the training dataset, of the model and its assessment can all have an impact on the trust experienced by the ML end users. It is therefore important to involve clinicians and patients early on in the process, to make sure that the developed models address existing gaps and serve to empower/augment its users with new capabilities or more time for other tasks of higher human added-value. The training dataset should also be representative of the local patient population, and any potential biases in it should be carefully investigated upfront, to avoid that the models learn to automate these biases, e.g. underdiagnosing a subset of the population.17 Models can also be more or less explainable. In the case of the linear regression model, the effect of each variable is easily extracted and understood from the fitted parameters. If the parameter is large, then the corresponding variable has more impact on the prediction. On the other hand, deep-learning models tend to behave as black-boxes where understanding the reason for the prediction is not always obvious, despite ongoing efforts in the data science community to address this limitation.18 Deep learning is therefore used a lot in image recognition, where ultimately the user/clinician can assess the result, whereas it tends to be less adopted in diagnosis/prognosis/risk predictions where an understanding of the underlying variables justifying the prediction are more needed. Tree-based algorithms (e.g. random forests, gradient boosting tree algorithms) are examples of opaque, yet somewhat explainable, models that have been used in recent nephrology articles.19,20
Perspectives – Digitization and AI in Nephrology in Switzerland, Where Do We Stand?
In Switzerland, digitization within the clinical realm is ongoing. Clinical laboratories have set up LIMS and EMR are being implemented throughout the country, fostered by initiatives like the Swiss Personalized Health Network ( www.sphn.ch ) and regulatory changes.21 A Swiss health record system is now available (DEP, dossier électronique du patient, www.patientendossier.ch/fr ). These incentives contribute to structuring the data and standardizing it with common terms and ontologies, which in turn can serve other purposes like data sharing and its reuse for research or for developing novel ML models. In nephrology in particular, the Swiss renal registry and quality assessment program ( www.swissnephrology.ch/srrqap/ ) and the Swiss Transplant Cohort Study ( www.stcs.ch ) are examples of how data that is structured and encoded with controlled vocabulary terms can be aggregated nationally, but also shared internationally with other similar registries like the European Renal Association/European Dialysis and Transplant Association (ERA/EDTA) registry ( www.era-online.org/en/ ) (Fig.1).
One step further, patient avatars are currently being developed. These digital or virtual SuperModels based on a patient-clinician-technician alliance integrate not only patients medical findings and history but also individual characteristics (risk factors, gene mutations, pharmacogenomics, environment, lifestyle behaviors, adherence to treatment recommendations and socioeconomics patterns) and diagnostic tools available on health records, mobile health systems and other platforms.22 During the Covid-19 pandemic situation, telemedicine also became an important issue, especially for patients taking immunosuppressive agents to avoid personal visits but also for patients performing home dialysis (home-hemodialysis or peritoneal dialysis). Video chats helped to stay connected and to resolve treatment or access problems. Industrial companies now also offer cloud-based solutions to monitor treatment data. Prescriptions can be done remotely (e.g. Baxter Sharesource https://renalcare.baxter.com/products/sharesource , TDMS therapy data management system www.freseniusmedicalcare.com ), thus contributing to the digitization of nephrology data.
The annual meeting of the Swiss Society of Nephrology in 2021 included a keynote on AI and digitization. The next steps would be to address clinical questions based on the already available rich data, identify additional data to be digitized and have the community engage even more into digitization and potential AI applications notably through events from medical societies.
Literature:
1 van Leeuwen KG et al.: Artificial intelligence in radiology: 100 commercially available products and their scientific evidence. Eur Radiol 2021; 31: 3797-04 2 Lemley KV: Machine learning comes to nephrology. J Am Soc Nephrol 2019; 30: 1780-81 3 Baxi V et al.: Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod Pathol 2021; 35: 23-32 4 Saez-Rodriguez J et al.: Big science and big data in nephrology. Kidney Int 2019; 95: 1326-37 5 Nagy M: Machine learning in oncology: what should clinicians know? JCO Clin Cancer Inform 2020; 4: 799-810 6 Provenzano M et al.: OMICS in chronic kidney disease: focus on prognosis and prediction. Int J Mol Sci 2021; 23: 336 7 El Naqa I, Murphy MJ (eds.): Machine and deep learning in oncology, medical physics and radiology. Cham: Springer; 2022 8 Bülow RD: How will artificial intelligence and bioinformatics change our understanding of IgA Nephropathy in the next decade? Semin Immunopathol 2021; 43: 739-52 9 Heerspink HJL et al.: Effect of dapagliflozin on the rate of decline in kidney function in patients with chronic kidney disease with and without type 2 diabetes: a prespecified analysis from the DAPA-CKD trial. Lancet Diabetes Endocrinol 2021; 9: 743-54 10 Rossing P et al.: Efficacy and safety of finerenone in patients with chronic kidney disease and type 2 diabetes by GLP-1RA treatment: A subgroup analysis from the FIDELIO-DKD trial. Diabetes Obes Metab 2022; 24: 125-34 11 Sirdah MM, Scott Reading N: Genetic predisposition in type 2 diabetes: A promising approach toward a personalized management of diabetes. Clinical Genetics 2020; 98: 525-47 12 Liu KD et al.: Initiative: recommendations for awareness, recognition, and management of AKI. Clin J Am Soc Nephrol 2020; 15: 1838-47 13 Chaudhary K et al.: Utilization of deep learning for subphenotype identification in sepsis-associated acute kidney injury. Clin J Am Soc Nephrol 2020; 15: 1557-65 14 Woo SH et al.: Development and validation of a web-based prediction model for AKI after surgery. Kidney360 2021; 2: 215-23 15 Seyahi N, Ozcan SG: Artificial intelligence and kidney transplantation. World J Transplant 2021; 11:277-89 16 Emmert-Streib F et al.: An introductory review of deep learning for prediction models with big data. Front Artif Intell 2020; 3: 4 17 Norori N et al.: Addressing bias in big data and AI for health care: a call for open science. Patterns 2021; 2: 100347 18 Belle V, Papantonis I: Principles and practice of explainable machine learning. Front Big Data 2021; 4: 688969 19 Flechet M et al.: AKIpredictor, an online prognostic calculator for acute kidney injury in adult critically ill patients: development, validation and comparison to serum neutrophil gelatinase-associated lipocalin. Intensive Care Med 2017; 43: 764-73 20 Segal Z et al.: Machine learning algorithm for early detection of end-stage renal disease. BMC Nephrol 2020; 21: 1-10 21 De Pietro C, Francetic I: E-health in Switzerland: the laborious adoption of the federal law on electronic health records (EHR) and health information exchange (HIE) networks. Health Policy 2018; 122: 69-74 22 Brown SA: Principles for developing patient avatars in precision and systems medicine. Front Genet 2015; 6: 365
Das könnte Sie auch interessieren:
Chloride in the spotlight – the hidden driver of acid-base homeostasis
Chloride is the most abundant extracellular anion and contributes fundamentally to osmolality, electroneutrality, and acid-base balance. Despite this central role, it has often received ...
Nierenkrebs – gegenwärtige Strategien und zukünftige Trends in der Therapie
Nichtklarzellige Nierenzellkarzinome (non-ccRCC) sollen zwar nach den gleichen Standards wie ccRCC behandelt werden, die Outcomes sind jedoch schlechter. Bei ccRCC hat sich die adjuvante ...
Prä- und postoperatives Transplantationsmanagement in der hausärztlichen Praxis
Hausärzte spielen in der Betreuung von Patienten vor und nach Nierentransplantation, aber auch bei der Begleitung von potenziellen Lebendspendern von der Auswahl bis zur Nachbetreuung ...